-
Biosketch
Education PhD (Aug 2011–Jun 2016): Computational Biology
Molecular Biophysics Unit
Indian Institute of Science
Thesis Title: Secondary Structures in Proteins: Identification and Analyses
Supervisor: Prof. Manju BansalB.Tech. (Aug 2005–May 2009): Bioinformatics
School of Chemical & Biotechnology
SASTRA UniversityWork Experience Assistant Professor (Oct 2024 - Present): Computational Protein Design
Indian Institute of Technology
PalakkadSenior Bioinformatics Person (Dec 2023 – Oct 2024): Computational Protein Design
T-Therapeutics Ltd.
Cambridge, UKResearch Associate (Jan 2017 – Apr 2023): Computational Protein Design
School of Chemistry
University of BristolProject Assistant (Jun 2009 – Jul 2011): Structural Bioinformatics
Molecular Biophysics Unit
Indian Institute of ScienceEditorial Activities Editorial Board Member: Scientific Reports -
Research
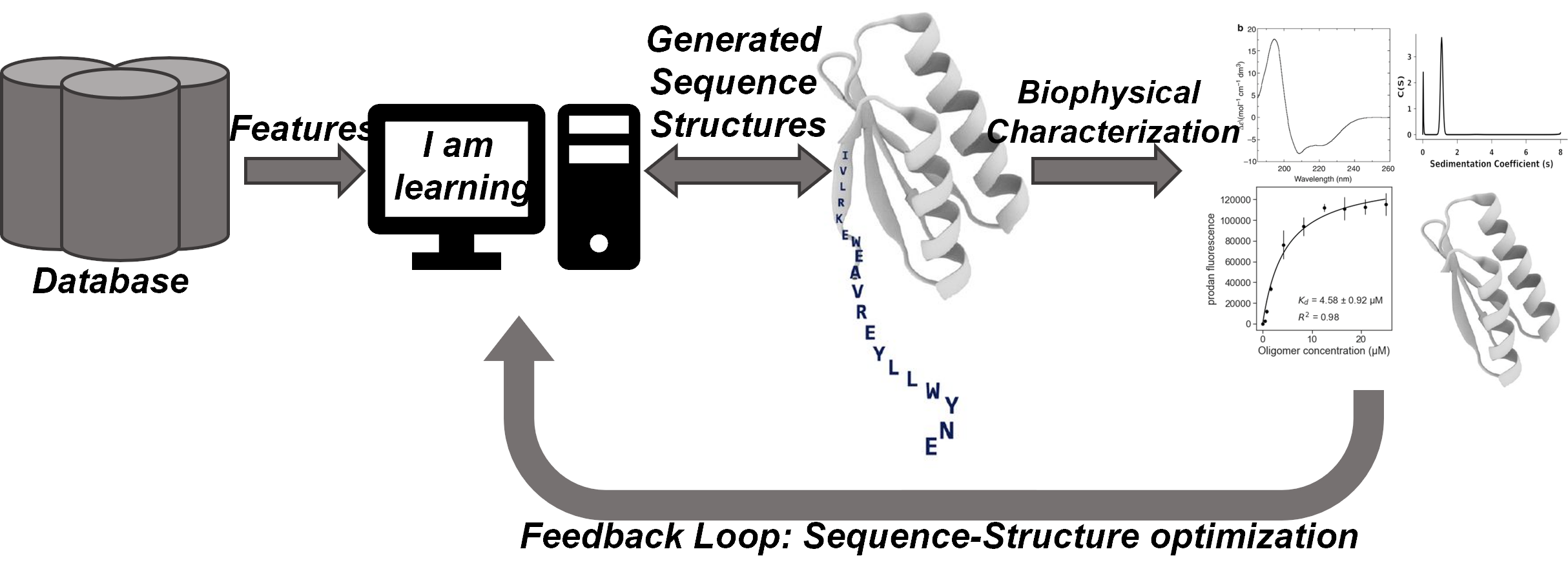
Figure 1: A schematic diagram for the proposed protein design approach.
-
Teaching
BS5002: Introduction to Bioinformatics (Elective)
BT2010: Life Sciences (B. Tech)
-
Additional Information
TitlePublicationsDescription
Research Publications in Journals
An atlas of protein homo-oligomerization across domains of life. H. Schweke, T. Levin, M. Pacesa, C. A. Goverde, P. Kumar, et al, (2023) Cell, 187, 999-1010.
CC+: A Searchable Database of Validated Coiled coils in PDB Structures and AlphaFold2 Models. P. Kumar*, R. Petrenas, W.M. Dawson, H. Schweke, E.D. Levy and D. N. Woolfson*, (2023) Protein Science, e4789.
De novo design of discrete, stable 310-helix peptide assemblies. P. Kumar*, N.G. Paterson, J. Clayden and D. N. Woolfson*, (2022) Nature, 607, 387–392.
Socket2: A Program for Locating, Visualising, and Analysing Coiled-coil Interfaces in Protein Structures. P. Kumar* and D. N. Woolfson*, (2021) Bioinformatics, 37, 23, 4575–4577.
RNAHelix: computational modeling of nucleic acid structures with Watson-Crick and non-canonical base pairs. D Bhattacharyya, S Halder, S Basu, D Mukherjee, P. Kumar and M.Bansal, (2017) Jl of Computer-Aided Molecular Design, 1–17.
Identification of structural and functional analyses of PolyProline-II helices in globular proteins. P. Kumar and M.Bansal, (2016) Jl of Structural Biology, 196, 3,414–425.
Dissecting π-helices: Sequence, Structure and Function. P. Kumar and M.Bansal, (2015) FEBS Journal, 282, 22, 4415–4432.
Identification of local variations within secondary structures of proteins. P. Kumar and M.Bansal, (2015) Acta Crystallographica Section D 71, 5, 1077–1086.
MolBridge: A program for identifying non-bonded interactions in small molecules and biomolecular structures. P. Kumar, S. Kailasam, S. Chakraborty and M. Bansal (2014) J Appl Crystallogr 47, 5, 1772–1776.
HELANAL-Plus: a web server for analysis of helix geometry in protein structures. P. Kumar and M.Bansal, (2012) J Biomol Struct Dyn., 30,773–783.
Identification, Activity and Disulfide connectivity of C-di-GMP Regulating proteins in M. tuberculosis. K. Gupta, P. Kumar and D. Chatterji, (2010) PLoS ONE 5, e15072
Chapters in Books
Biomolecular Structures: Prediction, Identification and Analyses. P. Kumar, S. Halder and M. Bansal in Reference Module in Life Sciences, 2019, Vol 3, pp. 504–534.
Defining α-helix geometry by Cα atom trace vs (φ-ψ) torsion angles: a comparative analysis. A. Shelar#, P. Kumar# and M.Bansal in 'Biomolecular Forms and Functions' (Ed. M. Bansal and N.Srinivasan) IISc, Bangalore, 2013, pp.116–127.
* corresponding author
# both authors contributed equally
INDIAN INSTITUTE OF TECHNOLOGY PALAKKAD
Kanjikode | Palakkad | Kerala - 678623
Kanjikode | Palakkad | Kerala - 678623
0491 209 2001 (Office) |
info@iitpkd.ac.in
Copyright ©2023 Indian Institute of Technology Palakkad. All Rights Reserved.


